1. Introduction
Campylaephora boydenii (E.S.Gepp) Barros-Barreto & Maggs (Ceramiaceae, Florideophyceae) is an endemic red algal species distributed in the Northwest Pacific region such as Korea, China, and Japan (Boo and Lee 1994; Guiry and Guiry 2023). All nine species of the genus Campylaephora J.Agardh were recognized with a full cortication of thallus, dichotomous and pseudodichotomous branchings with adventitious branches (Cho et al. 2001; Cho and Riosmena-Rodriguez 2008; Barros-Barreto et al. 2023). The DNA sequence-based taxonomy and phylogeny of species became common in the genus because of its morphological plasticity and wide range of distributions (Maggs et al. 2002; Won and Cho 2011). Some ceramiacean species used in population demography and inter-species diversity based on variable organelle markers (Yang et al. 2008, 2009; Bruce and Saunders 2015). Organellar genomes of the ceramialean species were successfully used in phylogeny and evolution studies (Lee et al. 2016; Díaz-Tapia et al. 2019), however, a few plastid and mitochondrial genomes are available in the tribe Ceramieae such as Campylaephora sungminbooi (J.R.Hughey & G.H.Boo) Barros-Barreto & Maggs (Hughey and Boo 2016), Centroceras clavulatum (C.Agardh) Montagne (Díaz-Tapia et al. 2019), Ceramothamnion japonicum (Okmura) M.J.Wynne & C.W.Schneider (Yang et al. 2015; Lee et al. 2016) and unidentified Gayliella sp. (MK814659; Díaz-Tapia et al. 2019). Thus new plastid genome of C. boydenii will provide significant information to increase available molecular markers and their applications in studies such as ecology, genetic diversity and phylogeny.
2. Materials and methods
In the present study, live materials were obtained from the collection site, Namsungri, Haenam, Jeonnam, Korea (34.321238 N, 126.601642 E) (Fig. 1A-1B) and thalli were transferred to the laboratory along with field waters. High-quality total DNA was extracted from live materials (ca. 1 g wet weight) cleaned by sterilized seawater using NucleoBond AXG 100 Kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany), and leftover thalli dried and kept as voucher specimen ECY.CB1905NSR.001. Total 10 individuals were collected, dried, and kept with silica-gel in the laboratory herbarium of the Korea Institute of Ocean Science & Technology (www.kiost.ac.kr, ecyang@kiost.ac.kr) under the voucher number (ECY.CB1905NSR.001-ECY.CB1905NSR.010). The identity of the specimen was confirmed by using standard sequencing of rbcL and BLAST searches (https://blast.ncbi. nlm.nih.gov). The quality and quantity of total DNA were tested by using the Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). The total quantity of DNA was 200 ng (19.2 ng/ul concentration) with purity ratios 260/280 = 1.77 and 260/230 = 1.57. The MGIEasy DNA Library Preparation Kit (MGI Tech, Shenzhen, China) was used for the NGS library construction and followed by whole genome sequencing using MGISEQ-2000 (LAS, Gimpo, Korea). From a total of 10 GB raw data, the plastid contig was assembled with the NOVOPlasty V.4.3.1 (Dierckxsens et al. 2017). Runs of perl script NOVOPlasty4.3.1.pl repeated two times using default configure options but different reference seeds, i.e., rbcL of C. boydenii MW366520 and DQ350389. Plastid genome was confirmed by aligning raw reads on assembly using the protocols for sequencing depth and coverage map with BWA V.0.7.17 and SAMtools V.1.16.1 (Ni et al. 2023). Annotation was done with the Geneious Prime V.2023.0.1 (www.geneious.com) assisted by a Ceramothamnion japonicum (NC_031174, Lee et al. 2016) and Campylaephora sungminbooi (NC_031211, Hughey and Boo 2016) as reference genomes, tRNAscan-SE online (Lowe and Chan 2016), and NCBI BLAST searches. The plastid genome (ptDNA) map drew in OGDRAW online (https://chlorobox. mpimp-golm.mpg.de/OGDraw.html, Greiner et al. 2019) and CPGView (Liu et al. 2023). The phylogeny of Campylaephora was constructed by using rbcL (total 1,467 bp). The maximum likelihood phylogeny and bootstrap (-# 1,000) analysis were performed with the GTR+G model and default settings (-f a) by using RAxML V.8.2.12 (Stamatakis 2014).

Fig. 1.
Images of Campylaephora boydenii (Ceamiaceae, Florideophyceae). The photographs were taken by Eun Chan Yang from intertidal zone of the Namsungri, Haenam, Jeonnam, Korea (34.321238 N, 126.601642 E) at 20th May 2019. (A) Thallus under water in tidal pool. Thalli are bright to rosy-red color, uniseriate filamentous, completely corticated and densely entangled on Sargassum sp. (B) Typical dichotomous branches and moderately inrolled apical tip of C. boydenii (Voucher ID: ECY.CB1905NSR.001) were found under the stereo microscope.
3. Results and discussion
The rbcL phylogeny of Campylaephora showed monophyly of the genus (91% bootstrap support; Fig. 2) and the closer relationship of C. boydenii to C. sungminbooi, C. gardneri (Kylin) Barros-Barreto & Maggs and unidentified Campylaephora sp. GQ179819 (64% bootstrap support). The C. boydenii ECY.CB1905NSR.001 collected from Namsungri (Haenam, Korea OQ731916) grouped with those from Korea (Anin AY945781, Hoidong AY770656, and Jindo EF613510) and Japan (Hakodate DQ350389 and Uranohama MW366520) (100% bootstrap support). The rbcL sequence identity in C. boydenii ranged from 99.68% (between Namsungri, Haenam, Korea and Uranohama, Japan MW366520) to 100% (between Anin, Gangwon, Korea AY945781 and Hakodate, Hokkaido, Japan DQ350389).
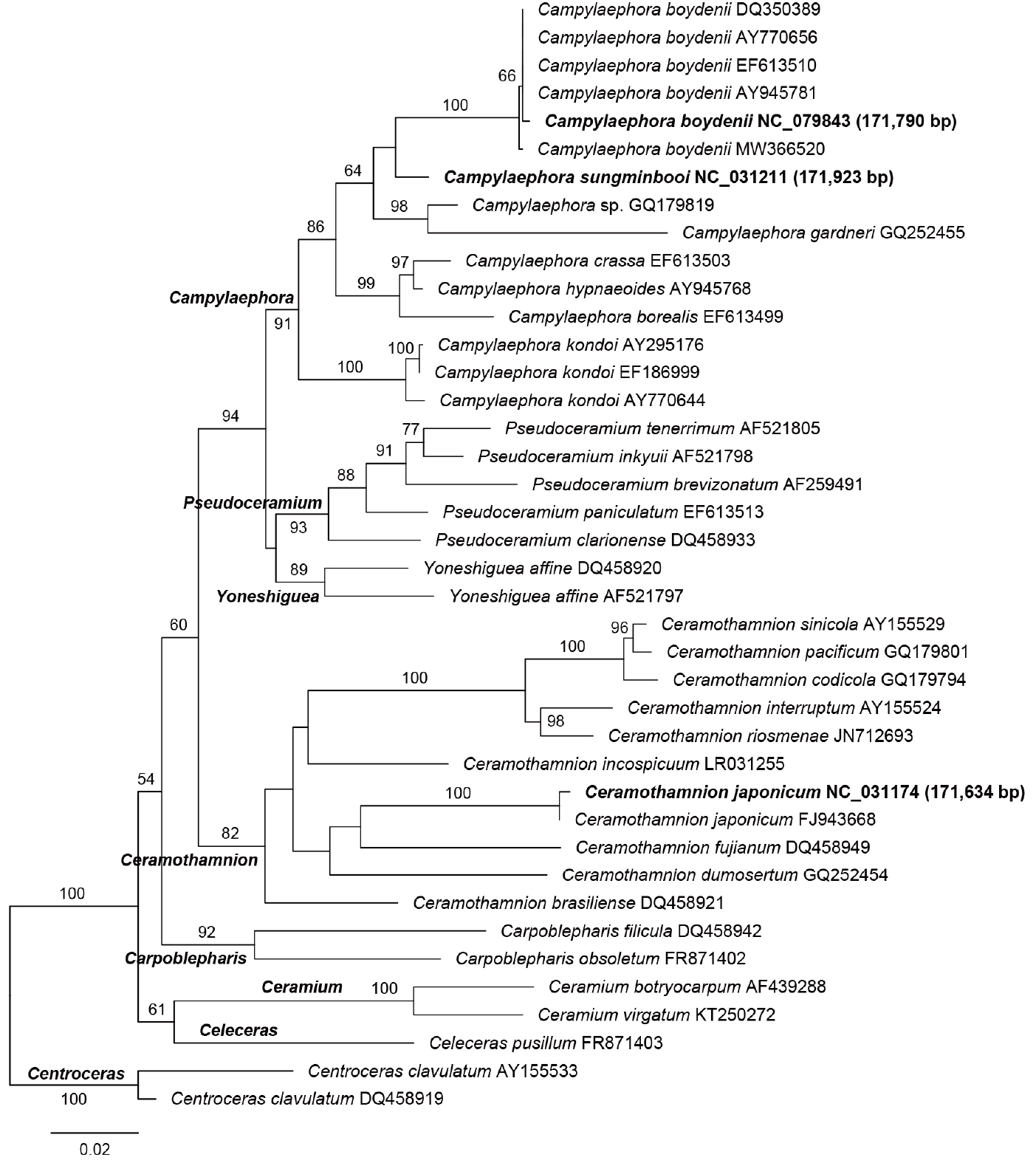
Fig. 2.
Phylogeny of the tribe Ceramieae including (Ceramiaceae, Florideophyceae) based on rbcL. Three species with plastid genomes are indicated in bold and followed by accession number and total length. A total 29 species included eight Campylaephora, two Carpoblepharis, one Celeceras, one Centeroceras, two Ceramium, 10 Ceramothamnion, five Pseudoceramium, two Yoneshiguea, species. The Centroceras is considered an outgroup because the genus was basal positioned in the recent study. The maximum likelihood (ML) phylogeny of reconstructed by using RAxML V.8.2.12 with GTR+G model and default settings. The monophyly of each node was supported by bootstrap values (>50%) from 1,000 replications under the same evolutionary model of the ML search. The following sequences were used: Campylaephora boydenii EF613510 (Cho et al. 2008b), C. boydenii AY945781 (Cho et al. 2008a), C. boydenii DQ350389 (Yang et al. 2008), C. boydenii AY770656 (Yang et al. 2008), C. boydenii OQ731916 (this study), C. boydenii MW366520 (Barros-Barreto et al. 2023), C. sungminbooi NC_031211 (Hughey and Boo 2016), Campylaephora sp. GQ179819 (Barros-Barreto et al. 2023), C. gardneri GQ252455 (unpublished), C. hypnaeoides AY945768 (Cho et al. 2008a), C. crassa EF613503 (Cho et al. 2008b), C. borealis EF613499 (Cho et al. 2008b), C. kondoi EF186999 (Yang et al. 2008), C. kondoi AY295176 (Yang and Boo 2004), C. kondoi AY770644 (Yang et al. 2008), Pseudoceramium inkyuii AF521798 (Cho et al. 2003a), P. tenerrimum AF521805 (Cho et al. 2003a), P. brevizonatum AF259491 (Lin et al. 2001), P. paniculatum EF613513 (Cho et al. 2008b), P. clarionense DQ458933 (De Barros-Barreto et al. 2006), Yoneshiguea affine DQ458920 (De Barros-Barreto et al. 2006), Y. affine AF521797 (Cho et al. 2003a), Ceramothamnion sinicola AY155529 (Cho et al. 2003b), C. pacificum GQ179801 (unpublished), C. codicola GQ179794 (unpublished), C. riosmenae JN712693 (Won and Cho 2011), C. interruptum AY155524 (Cho et al. 2003b), C. incospicuum LR031255 (unpublished), C. japonicum NC_031174 (Lee et al. 2016), C. japonicum FJ943668 (Yang et al. 2009), C. fujianum DQ458949 (De Barros-Barreto et al. 2006), C. dumosertum GQ252454 (unpublished), C. brasiliense DQ458921 (De Barros-Barreto et al. 2006), Carpoblepharis obsoletum FR871402 (Wolf et al. 2011), C. filicula DQ458942 (De Barros-Barreto et al. 2006), Ceramium botryocarpum AF439288 (Maggs et al. 2002), C. virgatum KT250272 (Bruce and Sanuders 2015), Celeceras pusillum FR871403 (Wolf et al. 2011), Centroceras clavulatum AY155533 (Cho et al. 2003b), C. clavulatum DQ458919 (De Barros-Barreto et al. 2006).
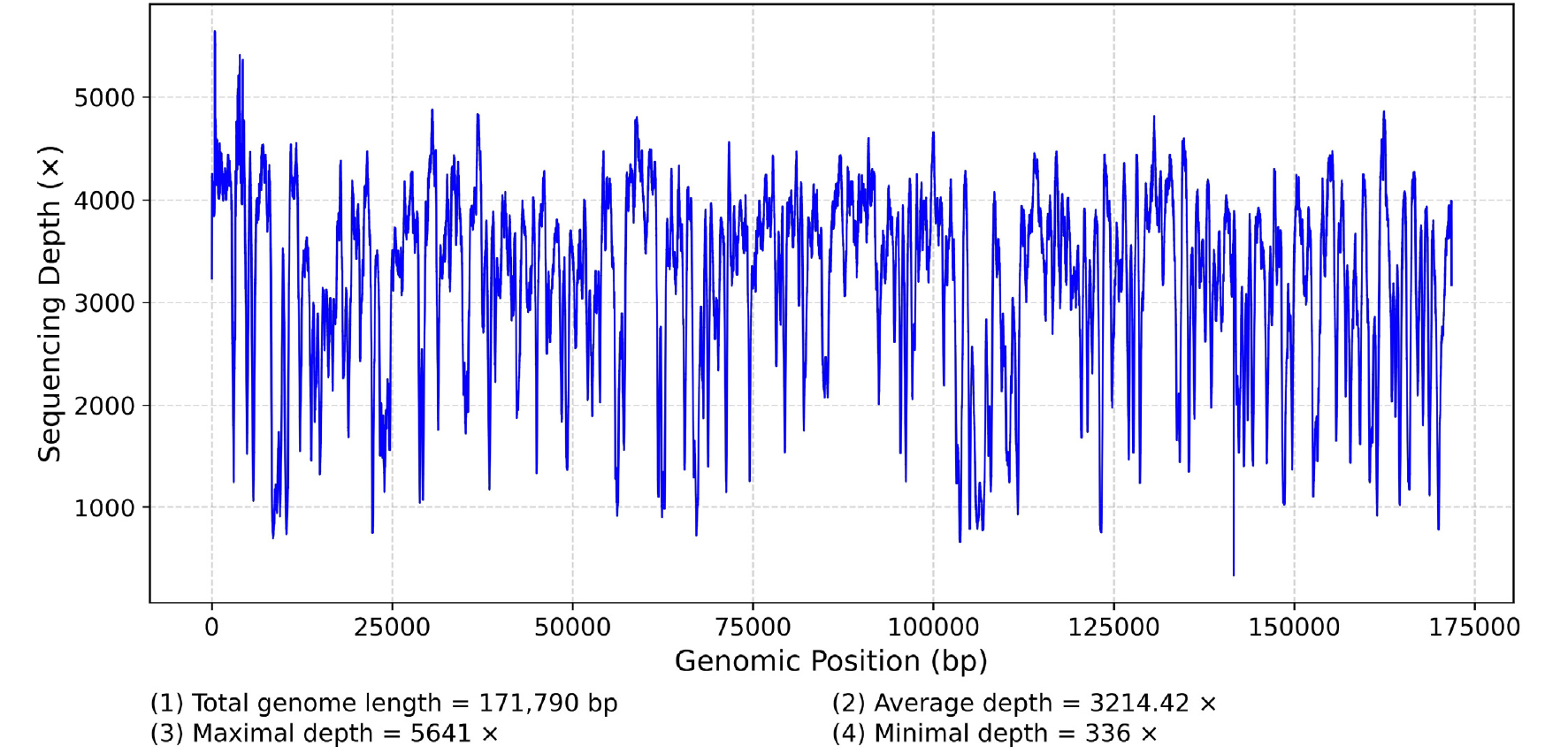
The average and minimum read mapping depths of the plastid assembly were 3,214 X and 336 X, respectively (Fig. S1). The complete plastid genome of Campylaephora boydenii (GenBank accession: OQ731916) was 171,790 bp in length with 27.7% GC content and 233 genes (154,497 bp; 89.8% of ptDNA; Table 1) including 200 protein-coding sequences (CDS), one group II intron, 28 transfer RNA (tRNA), three ribosomal RNA (rRNA), and one transfer-messenger RNA (tmRNA). Most of CDS’ codons started with ATG (182 out of 200 total CDSs). However, there were five alternative start codons found in the C. boydenii ptDNA, such as ATA for seven CDS (i.e., cpcG, fabH, infB, petP, rpl19, ycf38, and orf259), GTG for five CDS (atpF, chlI, psaF, rbcS, and trxA), TTG for three CDS (petM, ycf27, and ycf65), ATT for two CDS (ycf37 and ycf80), and ATC for argB.
Table 1.
Summary of plastid genomes contents in the Ceramiaceae.
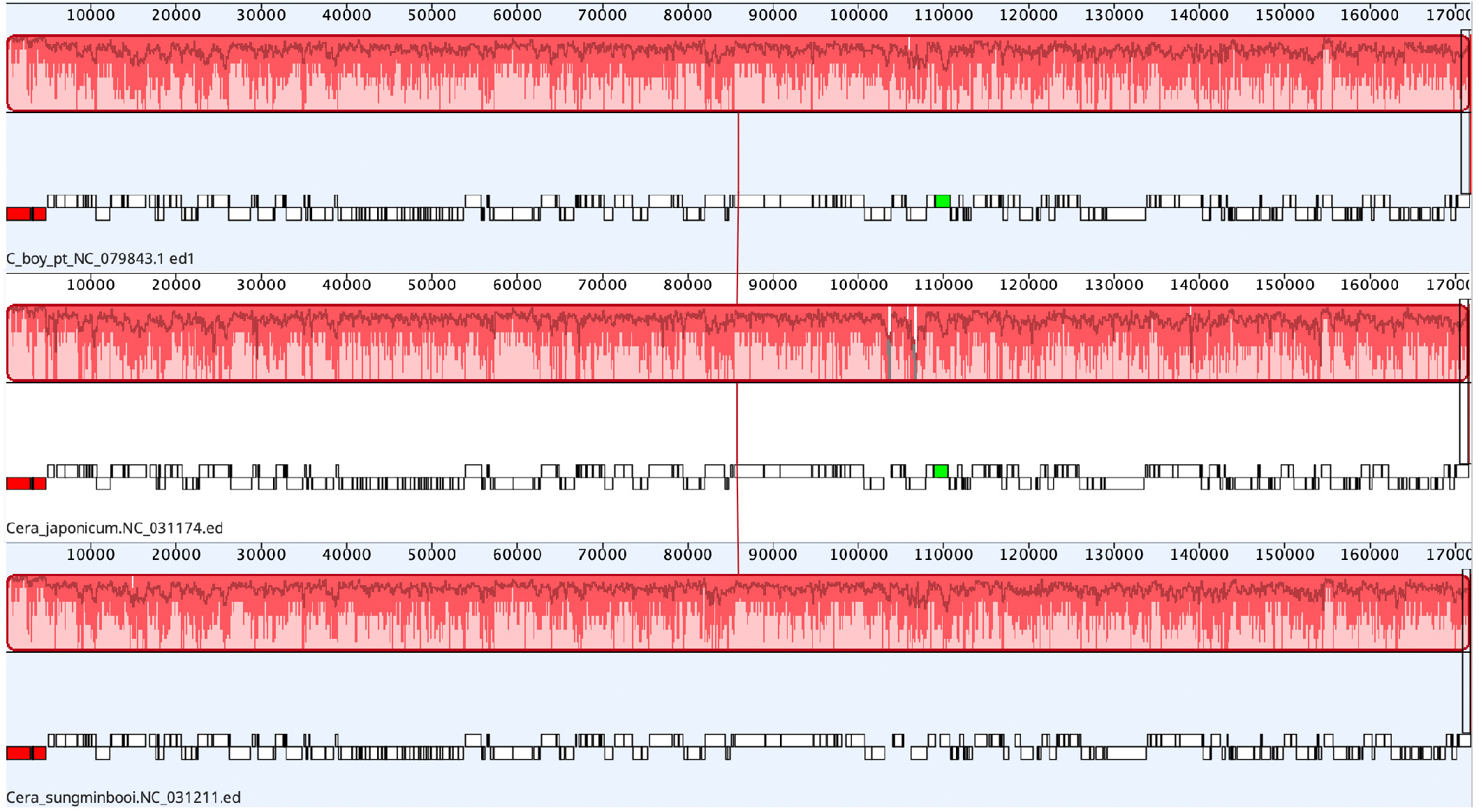
We compared the plastid genome of C. boydenii (Fig. 3) to those of C. japonicum (NC_031174, 171,634 bp with 27.9% GC; Lee et al. 2016) and C. sungminbooi (NC_031211, 171,923 bp with 27.7% GC; Hughey and Boo 2016) (Table 1). There was no syntenic variation (reversion or translocation) found among species in genome alignment (Fig. S2). The gene contents (233 genes) and synteny are highly conserved (86.2% pairwise identity) among three ceramiacean species. At DNA level, pairwise identities of C. boydeniii ptDNA are 93.3% to C. sungminbooi and 82.1% to C. japonicum (cf. 82.4% between those of C. japonicum and C. sungminbooi). Of 200 CDS, four CDS shows significant difference (± 5%) in length, i.e., cpcG, petP, rpl33, and thiS. The length of cpcG (encodes phycobilisome rod-core linker protein) of C. japonicum is 696 bp, however, those of C. boydenii and C. sungminbooi both are 768 bp. In petP (encodes cytochrome b6-f complex subunit), C. sungminbooi shows 204 bp in length, and the other two species show 177 bp. The length of rpl33 (encodes ribosomal protein L33) is 246 bp for C. sungminbooi and those are 204 bp for C. boydenii and C. japonicum. For the last, the full length of thiS (encodes thiamine biosynthesis protein S) are 216 bp for C. japonicum and 333 bp for C. boydenii and C. sungminbooi. Three ribosomal RNA (5S, 16S, and 23S rRNA) show no significant difference in both length and DNA sequence among three Campylaephora species. Average pairwise sequence identity of three plastids are 96.6% for 5S rRNA (length = 117 bp, 41.6% GC content), 97.4% for 16S rRNA (1,477 bp, 48.9% GC), and 97.3% for 23S rRNA (2,747 bp, 45.6% GC). In general, the GC content of all rRNAs (46.6% for C. boydenii, 46.3% for C. japonicum, and 49.6% for C. sungminbooi) shows higher than those of CDS (28.3% for both C. boydenii and C. sungminbooi, and 28.5% for C. japonicum). The trnM is a unique tRNA that contains a group II intron including with an intronic-ORF in all Campylaephora ptDNA. In both C. boydenii and C. sungminbooi, the trnM includes an intron and intronic-ORF (orf364, 1,098 bp in length). The trnM of C. japonicum includes an intron and intronic-ORF (orf357, 1,071 bp).

Fig. 3.
The plastid genome map of Campylaephora boydenii. Genes are color-coded by their functional classification. The colored boxes on the inside and outside of the circle are transcribed in a clockwise and counter-clockwise directions, respectively. A gray region of inner circle indicating the GC content.
Systematics and taxonomy of ceramialean red algae based on DNA (plastid marker, multi-genes, and genome) uncovered the genetic diversity of the group and morphological evolution of Rhodophyta (e.g., Zuccarello et al. 2002; Yang et al. 2008; Díaz-Tapia et al. 2017, 2019; Barros-Barreto et al. 2023). Despite of importance and usefulness of ceramialean ptDNA on genetic diversity, phylogeny, and evolution studies, however, a few plastids were reported as complete genomes (Hughey and Boo 2016; Lee et al. 2016; Díaz-Tapia et al. 2019). In order to increase our understanding of red algal genetic diversity in nature, additional complete plastid genomes will be required in the near future, such as Ceramium virgatum Roth, the type species of the family Ceramiaceae.